The problem* with ProteinGym
So just to start off, there is no problem with ProteinGym, and this post is not to hate on ProteinGym. ProteinGym is awesome. It was a super good idea, it is well-executed, and it’s truly a tremendous diversity of amazing datasets (each of which, of course, was itself a ton of work to produce), from binding affinities, to catalytic rate, to growth rate, and many other kinds of readouts.
And that diversity of readouts, I think is part of the problem. Not a problem with ProteinGym, but about how we use ProteinGym to evaluate the quality of our models. We treat it like a monolithic benchmark and report a single number, an average of performance across all the different assays. And they are all very different assays. Benchmarks for language models include design datasets where a natural protein is being engineered for novel function but provide the model no information about the desired ligand. Benchmarks include antibody binding data, growth data, thermostability data, as well as clinical data showing the macro-scale effects phenotypic effects of mutations.
In comparison to common NLP benchmarks, like HellaSwag, ProteinGym is a collection of nearly-unrelated tasks. In HellaSwag, we’re asking the model to look at a sentence and choose a completion from a list of them. In ProteinGym, we’re asking a model to make a prediction about a system where some crucial detail (such as what ligand is being measured, or the binding partner in an affinity assay) is often simply not provided, or cannot be provided in the case of the models we examine here. There’s no way to tell ESM to score the predicted activity of enzyme mutants in the context of a particular bound ligand. It’s a protein language model: it is never going to make the kinds of predictions you want for some arbitrary ligand you happen to be designing for. I’d like to examine the idea that using ProteinGym as a monolithic, single-number benchmark may be not be the best use for it, and think of some ways we can perhaps use it better.
Are there early-signal evals in ProteinGym?
HellaSwag does have some enviable properties that we’d like our eval datasets to have, one of which is early signal. Early signal is when you can see increasing progress on your benchmark as your model trains, or as your model sizes get larger, preferably early in the process, hence the name. Early signal is one important indicator of a quality benchmark.
Since the ProteinGym authors have already set up a fantastic benchmark suite and evaluated many different models on each of the collected datasets within ProteinGym, we have a lot of the information we need to answer this question. While it would be a bit out of scope for this blog post to retrain each model and keep track of its progress on ProteinGym as it trains (as you'd normally do when training a protein language model), we can at least look across model sizes. For the two types of models we'll consider (the encoder-only, BERT-like ESM-2 and the autoregressive ProGen and RITA), NLP research has shown that larger models trained on more data perform better. As model sizes increase, we should see eval performance increase, if the eval has sufficient range and signal to tell a good model from a bad one. Can we identify early signal evals in ProteinGym by looking at the performance across different model sizes in each of the three families?
To take a look at this, I inspected and assessed the Pearson correlation between model size and evaluation metric (Spearman rank correlation) for each of the datasets in ProteinGym. For each of the datasets in ProteinGym, I looked within each model family (ESM-2, RITA, and ProGen), basically looking for correlation. There is a lot of variation in the datasets when looked at this way, but some interesting trends emerge.
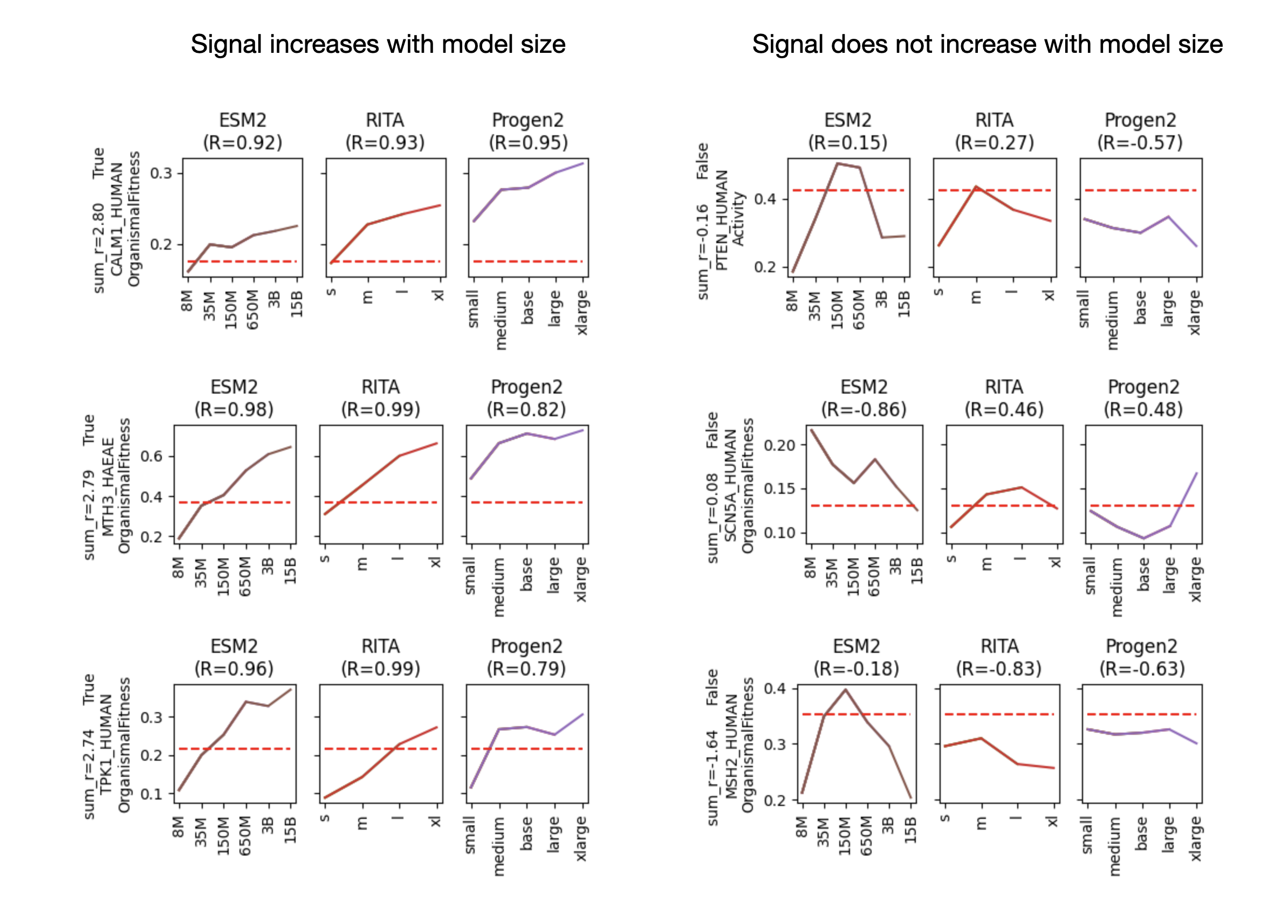
Figure 1. Examples of increasing (left panels) and not increasing (right panels) signal with model size in different datasets. In two sets of panels, we have three model families (ESM-2, RITA, and ProGen), evaluated on three datasets each. In each panel, the red dashed line indicates the performance of the EVmutation site-independent model evaluated on the dataset, a baseline model. The solid lines (brown for ESM-2, red for RITA, and purple for ProGen) trace the performance (measured by Spearman correlation) on the dataset for each of the model sizes. For the ESM-2 models, model sizes evaluated are 8M, 35M, 150M, 650M, 3B, and 15B param models. For the RITA models, model sizes are s, m, l, and xl. For the ProGen models, these are small, medium, base, large, and xlarge from the ProGen 2 series. The left panels are examples of datasets with a correlation between model size and prediction performance, and the right panels are examples of datasets where model performance is not correlated with size on the dataset.
While the examples in Figure 1 are specifically picked to be illustrative, there is a huge range in terms of this metric (correlation between model size and eval performance) across the different datasets. It could be that model performance on a subset of datasets may be a much better indicator of model performance than simply the mean of the Spearman correlations across all the different datasets.
Can we make better use of ProteinGym?
Again, ProteinGym is hella sweet. When using it as an eval for training models, there's already a diversity of ways of use the benchmark. When dealing with protein mutations, for example, how do you get a score from a transformer model like ESM? In the ESM-1v papers, the authors explore different ways of calculating a mutant score, and try them all on ProteinGym. The ProteinGym authors provide tons of scripts for running different models, as well.
As our desired and ability to generate more and more functional data on proteins grows, ProteinGym will also grow. I think we'll end with up a whole ProteinGym's worth of stability assay data, and a whole ProteinGym's worth of expression data, and a whole ProteinGym's worth of activity data, and that is going to be very cool. Until that time, I think we should be choosy about which datasets we include in our evals to get a sharper, clearer, and hopefully earlier signal about whether that new model we're trying out is any good. ProteinGym is a precious resource with a lot more subtlety than a single number, and we can use the depth and diversity of the datasets to our advantage.
Create your own evals for problems you care about
One of the most impactful things I did at Ginkgo was build task-specific evals for deep learning models, so we could determine which models worked well, and which worked poorly, on tasks that we cared about. For example, if you were designing for improved thermostability, you could look at the task-specific results in the evals and determine which model or set of models (or inference procedure) you should use. Because I built evals from real customer data, this was a rich resource for anyone designing proteins Ginkgo’s customers cared about.